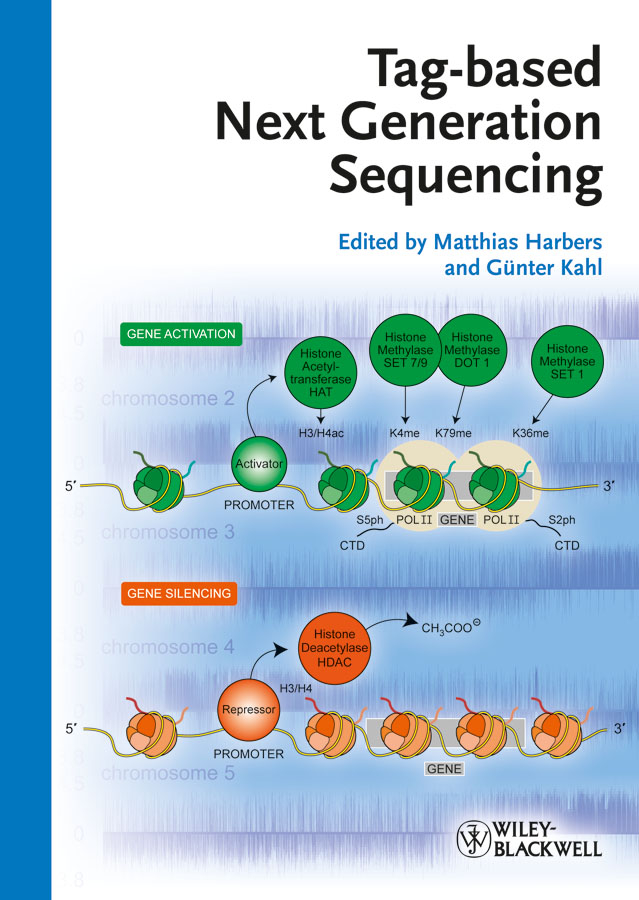
Tag-based approaches were originally designed to increase the throughput of capillary sequencing, where concatemers of short sequences were first used in expression profiling. New Next Generation Sequencing methods largely extended the use of tag-based approaches as the tag lengths perfectly match with the short read length of highly parallel sequencing reactions. Tag-based approaches will maintain their important role in life and biomedical science, because longer read lengths are often not required to obtain meaningful data for many applications. Whereas genome re-sequencing and de novo sequencing will benefit from ever more powerful sequencing methods, analytical applications can be performed by tag-based approaches, where the focus shifts from 'sequencing power' tobetter means of data analysis and visualization for common users. Today Next Generation Sequence data require powerful bioinformatics expertise that has tobe converted into easy-to-use data analysis tools. The book's intention is togive an overview on recently developed tag-based approaches along with means of their data analysis together with introductions to Next-Generation Sequencing Methods, protocols and user guides to be an entry for scientists to tag-based approaches for Next Generation Sequencing. INDICE: TAG-BASED NUCLEIC ACID ANALYSIS DeepSuperSAGE: High-throughput transcriptome sequencing with now- and next-generation sequencing technologies (Hideo Matsumura, Carlos Molina, Detlev H. Kruger, Ryohei Terauchi and Gunter Kahl) DeepCAGE: Genome-wide mapping of transcription start sites (Matthias Harbers, Mitch Dushay and Piero Carninci) Definition of promotome-transcriptome architecture using CAGEscan (Nicolas Bertin, Charles Plessy, Piero Carninci and Matthias Harbers) RACE: New applications of an old method to connect exons (Charles Plessy) RNA-PET: Full-Length transcript analysis using 5? and 3? paired-end-tag next generation sequencing (Xiaoan Ruan and Yijun Ruan) Stranded RNA-Seq: Strand specific shotgun sequencing of RNA (Alistair R.R. Forrest) Differential RNA sequencing (dRNA-seq): Deep-sequencing based analysis of primary transcriptomes (Anne Borries, Jorg Vogel and Cynthia M. Sharma) Identification and expression profiling of small RNA populations using high-throughput sequencing(Javier Armisen, W. Robert Shaw and Eric A. Miska) Genome-wide mapping of protein-DNA interactions by ChIP-seq (Joshua W.K. Ho, Artyom A. Alekseyenko, Mitzi I. Kuroda and Peter J. Park) Analysis of protein-RNA interactions with single-nucleotide resolution using iCLIP and next generation sequencing (Julian Konig, Nicholas J. McGlincy and Jernej Ule) Massively parallel tag sequencing unveils the complexity of marine protistan communities in oxygen-depleted habitats (Virginia Edgcomb and Thorsten Stoeck) Chromatin interaction analysis using paired-end-tag sequencing (ChIA-PET) (Xiaoan Ruan and Yijun Ruan) Tag-seq: Next-generation tag sequencing for gene expression profiling (Sorana Morrissy, Yongjun Zhao, Allen Delaney, Jennifer Asano, Noreen Dhalla, Irene Li, Helen McDonald, Pawan Pandoh, Anna-Liisa Prabhu, Angela Tam, Martin Hirst and Marco Marra) Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) (Paul G. Giresi and Jason D. Lieb) Identification of nucleotide variation in genomes using next generation sequencing (Hendrik-Jan Megens and Martien A.M. Groenen) Ditag genome scanning--a restriction-based paired-end sequencing approach for genome structrual analysis (Jun Chen, Yeong C. Kim and San Ming Wang) Next-generation sequencing of BAC clones for next-generation physical mapping (Robert Bogden, Keith Stormo, Jason Dobry, Amy Mraz, Quanzhou Tao, Michiel van Eijk, Jan van Oeveren, Marcel Prins, Jon Wittendorp and Mark van Haaren) HELP-tagging: Tag-based genome-wide cytosine methylation profiling (Masako Suzuki and John M. Greally) Second-generation sequencing library preparation: In vitro tagmentation viatransposome insertion (Fraz Syed) NEXT-GENERATION TAG-BASED SEQUENCING Movingtowards third-generation sequencing technologies (Karolina Janitz and Michal Janitz) Beyond tags to full-length transcripts (Mohammed Mohiuddin, Stephen Hutchison and Thomas Jarvie) Helicos single-mol
- ISBN: 978-3-527-32819-2
- Editorial: Wiley-VCH
- Encuadernacion: Cartoné
- Páginas: 606
- Fecha Publicación: 07/12/2011
- Nº Volúmenes: 1
- Idioma: Inglés
